

In this part of the tutorial, some basic knowledge of gromacs commands is assumed and not all commands will be given explicitly. The last step can be done with the tools available in the gromacs package and/or with ad hoc scripts. However, if you want to be sure you use the same version as the files provided as an example, download this version of martinize.py. Two first steps are done using the publicly available martinize.py script, of which the latest version can be downloaded from Github. solvate the protein in the wanted environment. generating a suitable Martini topology ģ. converting an atomistic protein structure into a coarse-grained model Ģ. Setting up a coarse-grained protein simulation consists basically of three steps:ġ.

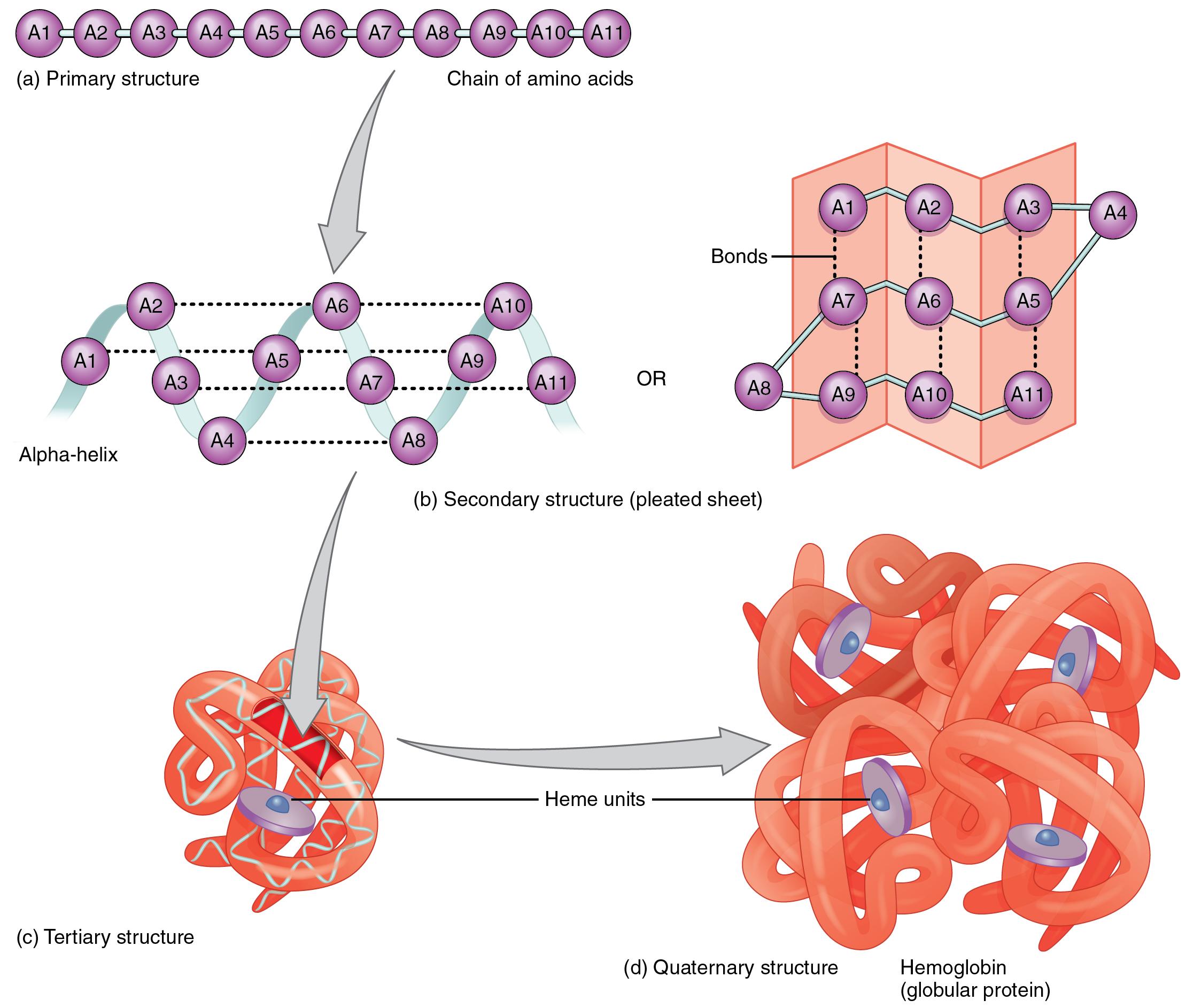
Conformational changes that involve changes in the secondary structure are therefore beyond the scope of Martini coarse-grained proteins. It is noteworthy that, even though the protein is free to change its tertiary arrangement, local secondary structure is predefi ned and thus imposed throughout a simulation. The secondary structure of the protein influences both the selected bead types and bond/angle/dihedral parameters of each residue. Each residue has one backbone bead and zero to four side-chain beads depending on the residue type.
Keeping in line with the overall Martini philosophy, the coarse-grained protein model groups 4 heavy atoms together in one coarse-grain bead. Tools and scripts used in this tutorial Soluble protein Soluble protein: the elastic network approach Rosetta homology modeling and ab initio fragment assembly with Ginzu domain predictionĬombination of ab initio folding and homology methodsĭetection of templates, alignment, modeling incl.(if you just landed here, we recommend you follow the tutorial about lipid bilayers first) Summary Proprietary platform, supported on Windows, Linux and Mac excluded ligand volumes), loop modelling, rotamer libraries for sidechain conformations, relaxation using MM forcefields. Template identification, use of multiple templates and accounting for other environments (e.g. Standalone program mainly in Fortran and Perl Satisfaction of contact and distance restraints Standalone program mainly in Fortran and Python Webserver with job manager, automatically updated fold library, genome searching and other facilities Remote template detection, alignment, 3D modeling, multi-templates, ab initio Template detection, alignment, 3D modeling Wraps external programs into automated workflowīLAST search, T-Coffee alignment, and MODELLER construction Remote homology detection, protein 3D modeling, binding site predictionĪutomated webserver and Downloadable program A unified interface for: Tertiary structure prediction/3D modelling, 3D model quality assessment, Intrinsic disorder prediction, Domain prediction, Prediction of protein-ligand binding residuesĪutomated webserver and some downloadable programs


 0 kommentar(er)
0 kommentar(er)
